FAIR knowledge graphs are a gold mine for AI. This article will explain why and provide examples how the knowledge graphs that power KnetMiner can be used by data scientists and developers.
The benefits of FAIR data
Just like many other future scientists and geeks, when I was a child, I liked to disassemble things around the home, understand what was inside, change the pieces around, build new things and amongst a family of mechanicians and farmers I had many opportunities to do so.
Now in 2020, after many years with Commodore computers and open source activism, I like to think of data behind articles, presentations or scientific software in a similar way: these digital artefacts were built on top of data to serve great purposes, such as easing data explorations with nice interfaces, exposing some scientific evidence or telling the public some knowledge that is worthwhile, or simply beautiful, to know.
However, these underlying data have much more potential, it’s like disassembling the LEGO castle and then building thousands of new things, many of which were never even thought of by those who wrote those leaflets included in the brick bucket. Indeed, there is now an entire world working on ways to publish and share bare data of all kinds, and in forms that make their reuse, their ‘reassembly’ as seamless as possible.
This is the open data world, which, among many fields, is contributing much to the world of science too.
In the (not always necessarily…) more complicated adult world, it isn’t always as easy as with toys, so the movement for open data needs to set principles, best practices and tools to make things working. If you’re reading this text, probably you already know that the main principles that currently help with publishing data in a good way are summed up by the clever FAIR acronym: in order to be useful in the wide, data should be
- easy to Find and Access
- Interoperable, so that integrating multiple data sets and applications is seamless…
- ..and makes data Reusable.
Our History of Knowledge Graphs
At KnetMiner, we are participating in this movement. Traditionally, KnetMiner data have been based on the Ondex OXL format, which, in practice, means knowledge graphs, a kind of data (and knowledge representation) that is great for integrating heterogeneous and complex information, typical for molecular biology data.
In 2018, we decided to push this forward. A pretty natural move, given that we were already integrating a number of publicly well-known data sources, and we were doing so by unifying them into a common model, a common format and a common vocabulary, all of which were the necessary basis to give our tools a common “understanding” of items like genes, proteins and phenotypes. So, we had some key aspects to produce FAIR data, by means of internal interoperability and common data representations, but providing truly FAIR knowledge graphs required further engineering and data modelling.
The KnetGraph project is all about our effort to provide FAIR knowledge graphs by adoption and integration of public, world-wide standards and offering integrated data via standardised endpoints and APIs.
Meet the KnetGraph Data Endpoints
To introduce how we are applying these ideas in practice, we have prepared a demo using Jupyter. If you haven’t come across Jupyter yet, it is an increasingly popular tool, used to edit and publish “live notebooks”. These are (web) documents where you can mix static text, programme fragments, graphics and more. The programmes you write there are typically short scripts, which do some very specific data analysis steps, such as fetching a CSV table and computing an average, or even drawing a chart!
Let’s see how this works by fetching some KnetGraph data from Jupyter:
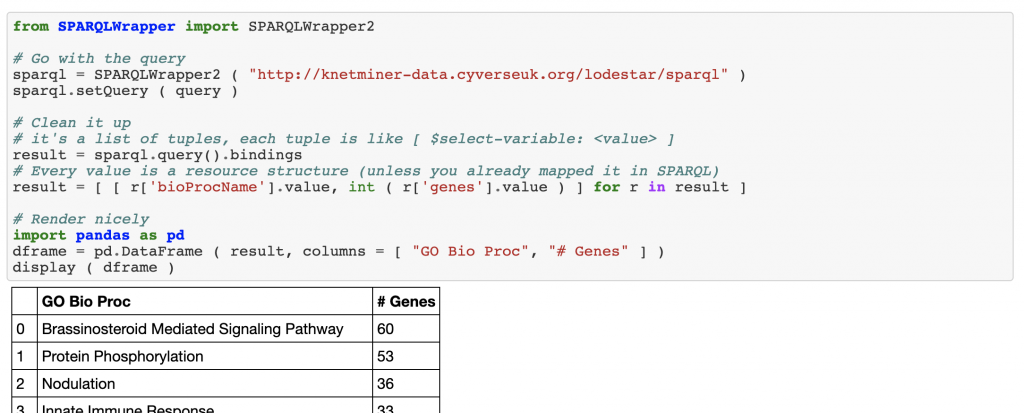
This is accessible live, here. As you can see, we’re building a narrative in a Jupyter notebook, where we show how we can perform a simple exploratory analysis. The example shows how to search for “yellow rust” in publications of the wheat knowledge graph, then, for genes linked to those publications, we generate statistics about the biological processes most commonly associated to those genes.
Standardising Data through the Web
Let’s talk about what’s going on conceptually: linked data and the Semantic Web are standards designed to leverage the web technology to share data online. They offer an entire stack of standardised tools and languages to address the problem of interoperability (and other FAIR principles). The main idea is that one can describe data through a common data syntax, the RDF, which is based on linked graphs, like in this example:
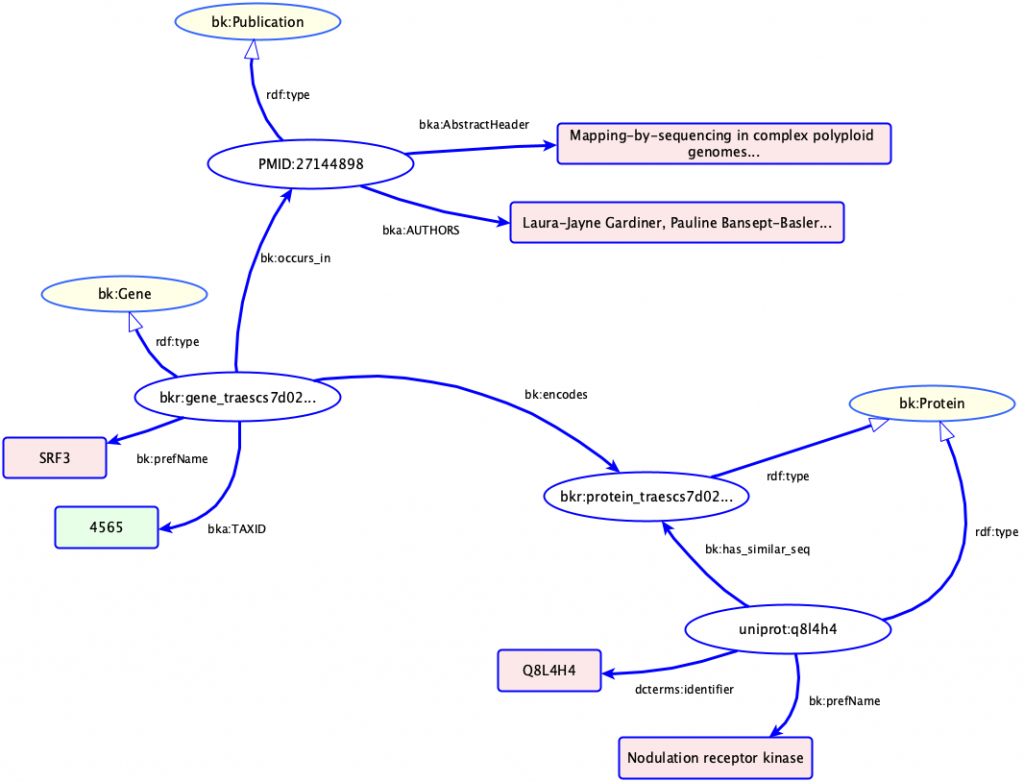
Above the syntax, you can define common data schemas and models. For instance, the figure above represents an occurrence (ie, a mention) of the gene SRF3 in a publication, along with the protein that gene encodes and another protein with a similar sequence.
The RDF data schemas are based on the RDF-compatible language OWL, which allows for the definition of very advanced data models that are named ontologies.
In the figure above, “Publication”, “Gene”, “Protein”, “identifier” and “encodes” are all examples of it. But don’t be afraid: recently the life science community has been using OWL to define simplified models, where the lack of advanced features are traded with simplicity. One example that we use a lot is bioschemas, a model that allows for dealing with common entities like genes, proteins, experiments, PubMed papers.
We have defined an OWL schema for KnetGraph (In the figure above, that is from where the prefixes like bk:, bka: come.), which also aspires to similar simplicity principles, and it’s mapped on standards like bioschemas (more later). So, the graph you see above is based on that schema. If you’re already familiar with KnetMiner terms and visualisations, you may have noticed that graphs like the one hereby are pretty similar. In the Jupyter notebook, you’ll find a SPARQL query. SPARQL is the Semantic Web component that plays the role of standardised query language. As you might expect, SPARQL queries reference the underlying data schemas (or, we should say ontologies in the more advanced cases) all the time.
To summarise, data can be modelled/schematised via OWL (which is encoded via RDF), we have a KnetGraph model/schema based on our own OWL schema, such schema is used to store KnetGraph RDF data into an RDF database (a so-called triple store) and this can be queried via standard web interfaces (both programmatically and user-dedicated), by means of SPARQL queries.
You can try that by copying-pasting the Jupyter example above. You could even do things like invoking SPARQL and fetch KnetGraph data from your own applications. Libraries like rdflib for Python or Jena for Java work well for that purpose.
Before starting coding, you might want to explore our RDF data by means of our SPARQL endpoint, which is based on the great LODEStar browser.
The Best of Two FAIR Worlds
But there’s more: have a look at this other Jupyter notebook:
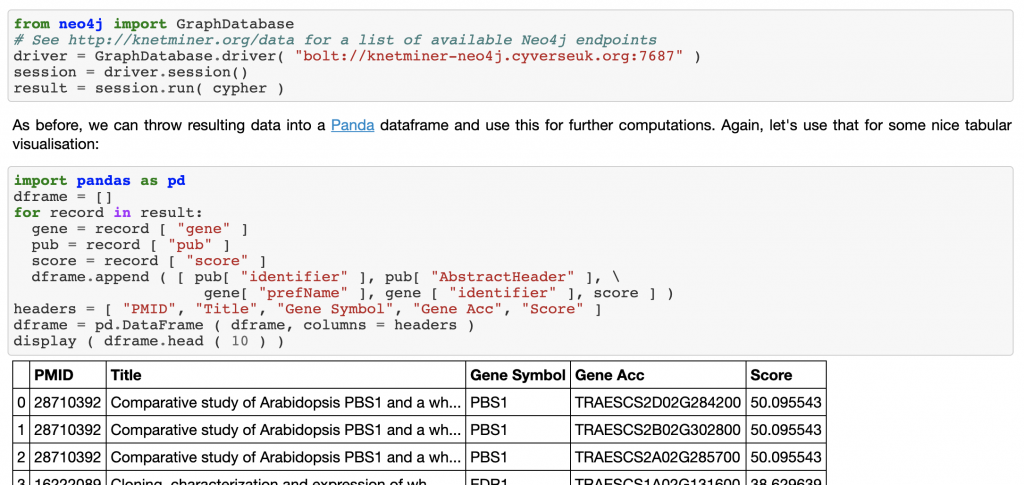
In this second example, we drill down through the results found via SPARQL, to explore the proteins and other entities associated with the brassinosteroid signaling pathway, which was the result most commonly associated with yellow rust.
Did you notice? We aren’t using SPARQL anymore, we have now switched to our Neo4j alternative. When we started the KnetGraph data standardisation and publishing effort, we soon realised that the linked data world and more recent technologies dedicated to knowledge graphs have sets of complementary advantages.
Comparing approaches, on the one hand, RDF and Semantic Web standards are great as common languages, they favour interoperability and standardisation (not least, because many standard schemas and ontologies already exist, especially in the life science world) and this is all good for realising a FAIR world. Plus, SPARQL is particularly good with tasks like complex graphs with many branches and other interconnections. We have discussed the details at the SWAT4LS 2018 workshop.
On the other hand, graph databases like Neo4j are often easier both to set up and to install. Their query language, Cypher, an emerging de-facto standard, is very compact and very good at expressing queries about chains of relations in a graph. Plus, there is a very nice end-user browser for Neo4j, which we offer on top of wheat, arabidopsis, and, our most recently developed, COVID-19 knowledge graph.
The SPARQL and Neo4j endpoints are aligned (and both are aligned to KnetMiner web instances): we base that on a common RDF/OWL representation of our data, based on the ontology mentioned above. Thanks to that, the schema that you can query in Cypher is conceptually the same that is available from the SPARQL endpoint (and it’s the same as that found in the KnetMiner web application). This allows you to either choose one endpoint or another, or even mix them together (though be aware that this requires skills on both…).
As you can see in the Jupyter notebook above, accessing Cypher or SPARQL endpoints from your applications is pretty simple: both are shipped with drivers for Python, Java and R, the query results can be easily accessed as objects defined in the respective programming language. If you’re already familiar with fetching data from SQL databases, you’ll reckon a similar approach.
Powering KnetMiner searches with semantic motifs
You can find a real application of the Cypher/Neo4j endpoint within KnetMiner: the Semantic Motif Graph traverser. KnetMiner’s Gene Rank algorithm (see Hassani-Pak et al., 2020) relies on biologically plausible paths that connect genes to other entities in the knowledge graph. For example, a graph path could link genes to proteins and to publications that mention such proteins. KnetMiner searches the knowledge graph for millions of paths that encode interesting and useful biological information. Since KnetMiner 4.0, developers or data curators can use Cypher to define the graph queries for KnetMiner. This offers more expressiveness and flexibility than our former simpler ad-hoc syntax.
There is more: Agrischemas!
The KnetGraph schema/ontology that we have shown so far is rather specific to KnetMiner. Many names that we use have primary definitions that are “in-house”. It doesn’t mean our data aren’t interoperable, though: we have mappings between our schema and other schemas and ontologies, such as bioschemas, schema.org, Dublin Core, SIO, SKOS. Moreover, many biological entities are annotated with OBO and other life science ontologies, such as Gene Ontology.
This facilitates tasks like combining KnetMiner data and other data sources modelled after the same schemas or annotated with the same ontology terms. For instance, we have annotations that make it possible to search for pathways imported from the BioPAX format and annotated with Gene Ontology terms.
We are also working to go beyond that! As part of the BBSRC funded Designing Future Wheat program, we are integrating various types of agriculture data related to plant biology on which KnetMiner is focused on, such as field trials and long-term experiments, gene expression data points, host-pathogen interactions. Moreover, we are defining new schemas, that we call agri-schemas, which reuse the already-mentioned bioschemas as much as possible. The idea is to base these data on the direct use of standardised schemas and to cover a wide variety of agricultural data. We will describe this in detail in a future post.
I’m a programmer, where can I go next?
If you are a data scientist or software developer, KnetGraph data and its graph query endpoints are yet another resource you can take advantage of, by flexibly querying our heterogeneous data, using them together with your own data or other biological and plant biology data. Adopting existing schemas and biological ontologies for your use cases helps this kind of task. We can help you building new applications powered by our FAIR knowledge graphs, endpoints and APIs, and we always welcome contributions to our open-source projects..
Starting points to know more are:
- All the schemas we mentioned above
- the sample queries available in the SPARQL browser
- the queries we use as semantic motifs (example)
- the queries we used for benchmarking RDF and Neo4j databases
If you want to learn more about the linked data and Neo4j technologies that we have presented, there is a vast amount of material, example 1, example 2.
And of course, you can get in touch with us!
And should I care if I’m not a programmer?
Yes, we might usefully work together even if you aren’t a geek! If you are a data manager and your organisation has KnetMiner-related data that would be worth sharing, either publicly, or internally, with collaborators or even customers, it would be ideal to model them using the same principles and standards that we presented in this article. In particular, some little modelling like the one we are applying to our own data, could go a long way in making your data more usable (for yourself and others) and visible. So, if you feel you might be in this situation, get in touch with us!
Acknowledgements
This work is funded by the UK BBSRC Designing Future Wheat (DFW) project. We are also grateful to the Robert Davey’s group and CyVerse UK, for providing the VMs that host our public KnetGraphs and helping with their set up and maintenance. Image sources: tree root network.










