Pilot study with VLPB
A consortium of European plant breeding companies (VLPB) have evaluated the value of KnetMiner to their businesses. Working together with the team at Rothamsted Research, the proof of concept project included virtual workshops and knowledge exchange events on using KnetMiner for data integration, knowledge mining and gene discovery.
Customising knowledge graphs with private data
The first aim of the POC was to assess the integration of confidential data into the rich knowledge graphs (KG) licensed by Rothamsted Research. VLPB partners were interested to customise KnetMiner knowledge graphs with proprietary genomics and trait datasets. “We tested the KnetBuilder tool for transforming our tabular data into semantic graphs and connecting it with the wealth of public knowledge stored in the KnetMiner KG”, said one of the VLPB partners.
We developed a special KnetBuilder tutorial with example knowledge graphs to accelerate deployment and customisation by our plant breeding partners allowing them to add custom data independently. “It is a big plus that we can run the data integration on our own servers where the private data is located”, commented a VLPB partner.
Deploying KnetMiner in the cloud
The second goal of the POC was to evaluate the deployment of KnetMiner in the cloud. As with many organisations, breeding companies use a mixture of cloud and on-premise computing infrastructures. VLPB partners tested the parallel deployment of tomato, pepper, potato and solanum (containing the 3 crops) datasets in Azure, AWS clouds and Kubernetes on-premise using our Docker images and scripts.
The memory requirements and app performance of KnetMiner were subsequently tested. KnetMiner presently uses an in-memory graph implementation to deliver ultra fast in-app searches. The memory footprint increases with the size of the KG and the number of graph queries. “We are moving towards a new architecture using RDF and Neo4j graph databases to standardise our KGs while at the same time reducing the memory requirements.”, said Keywan Hassani-Pak (Founder of KnetMiner). “I am very excited about our roadmap to build the next generation KnetMiner which will enable us and our customers to scale to more crops, insects and pathogens, integrate larger and wider datasets, improve interoperability and save cloud costs. Get in touch with us to hear more about co-development opportunities.”
Value to knowledge mining and gene discovery
To explore the added value of KnetMiner to gene discovery and crop improvement, VLPB members used the web application to search for a variety of traits, QTL, and candidate genes. “Researchers were able to search the database and find causal genes in the top 10 results of queries, which previously took them months to identify by creating mapping populations” said one of the VLPB partners. Feedback on KnetMiner from end users were largely positive with users commenting: “Having a KnetMiner where all public and private relationships for various crops are integrated would provide the end user a single place to ask more complex questions and get answers much faster than currently possible with unintegrated data.”
Scientists from a potato breeding company were excited about the richness of information available in the knowledge graph. “We especially liked the integration of different biological knowledge to discover gene-trait relations and the ranking of genes for traits of interest.” Other elements of the KnetMiner UI that caught users attention were the interactive network viewer KnetMaps and the new KnetSpace platform for scientists to manage and share gene knowledge networks.
An example gene network for PSY in tomato, potato and pepper along with model species information, trait associations and relevant publications has been made public in KnetSpace and can be explored here:
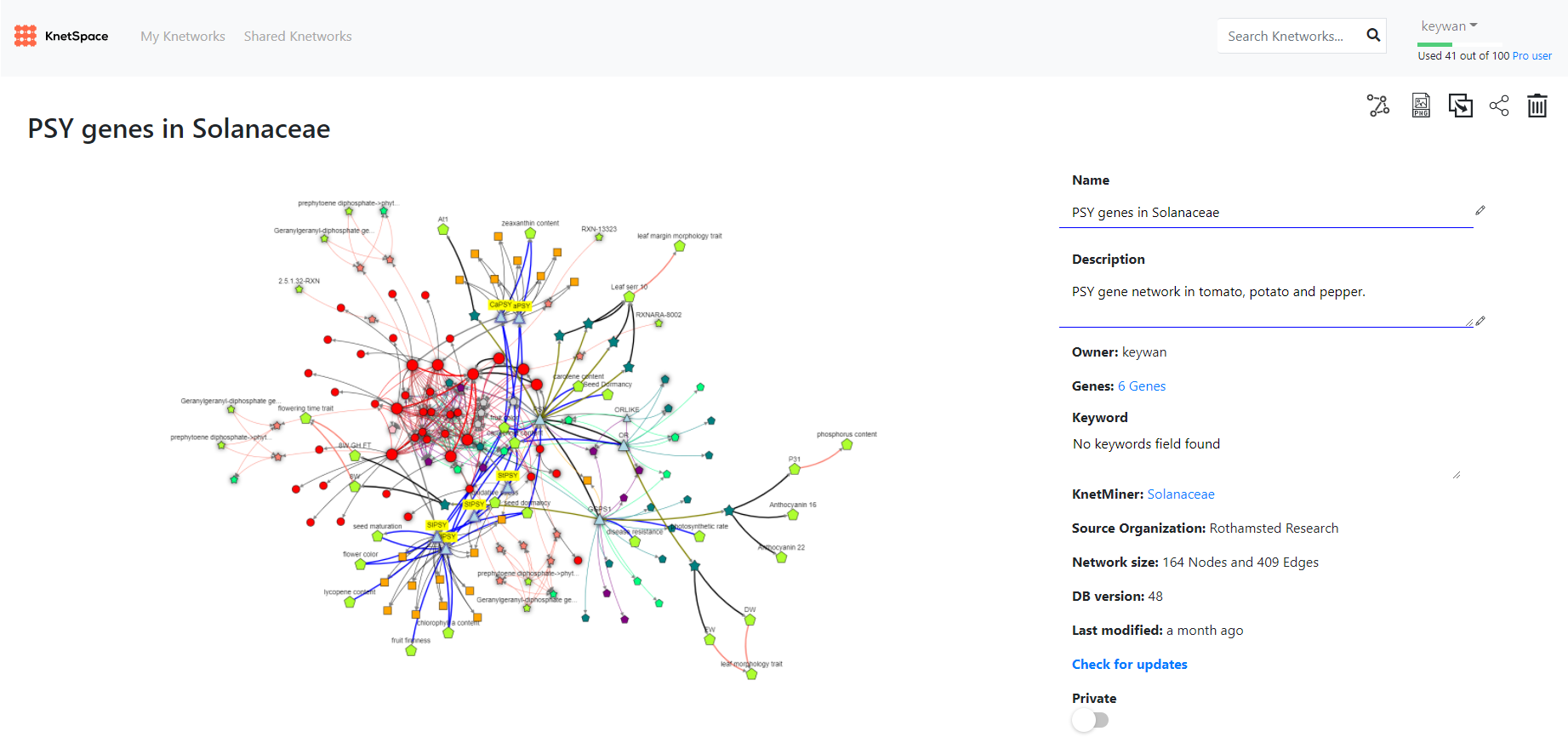
We see the real value of KnetMiner being in the areas of multi-dimensional fusion and global optimisation where, by comparison, a human cannot hold that much data depth or breadth together and in balance. An expert scientist might read and understand a single journal article better and follow a single thread or conclusion accurately but it is the integration and balance of the entirety of the indexed knowledge graph that cannot be beaten.
The verdict
The KnetMiner POC with VLPB has been an enormous success. The breeding companies have learned about the power of KnetMiner to accelerate gene discovery using an integrated and cross-species approach. We have learned about the requirements of different breeding companies and how they wish to further optimise KnetMiner to better fit their business environments. KnetMiner 5.0 will incorporate several new features that were requested as part of the POC to simplify on-premise deployment and customisation.
Interested to start a KnetMiner pilot study? Connect with us! We are fully committed to amaze your organisation and earn your business for the long run.










