Beyond Fluency: Why Biological Reasoning Requires Graphs, Not Just Prose
Why your LLM is a great writer but a poor biologist.
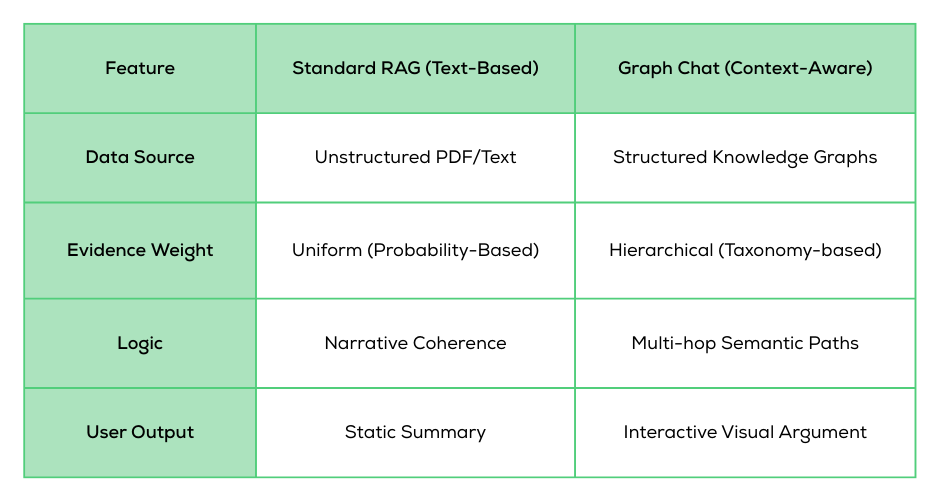
The current wave of “chat with your data” tools operates on a seductive but flawed premise: that scientific understanding can be reconstructed from text alone.
In the standard RAG (Retrieval-Augmented Generation) pipeline, documents are shredded into chunks, retrieved via similarity search, and fed into a Large Language Model (LLM). This works beautifully for summarization. However, it fails fundamentally for biological reasoning, where meaning is encoded in structured relationships, evidence hierarchies, and experimental context—not just prose.
The Limits of Flattening Science
Whether using GPT-5-class frontier models or specialized open-weight models like LLaMA and Mistral, a core limitation remains: Without explicit structure, LLMs infer biological relationships probabilistically.
As context windows expand into the millions of tokens, models are becoming better at remembering text, but not necessarily better at understanding biological evidence. Fine-tuning isn't the silver bullet, either. While it improves domain fluency, it introduces rigidity and overhead without teaching the model how to reason over live, structured data. In practice, fine-tuned models still tend to flatten evidence strength, smooth over contradictions, and prioritize narrative coherence over scientific accuracy.
Graph Chat: Context Engineering Over Fine-Tuning
We believe the path forward isn't "bigger models," but better context. Graph Chat replaces the flat text-chunking approach with precise context engineering.
Instead of searching for keywords, the system queries a large-scale biological knowledge graph to generate a specific sub-graph relevant to the user's query. This sub-graph captures the essential interactions, homologies, and trait associations that text-only models miss.
The "Secret Sauce": An Evidence Taxonomy
To bridge the gap between a "graph" and "reasoning," we developed a formal schema and evidence taxonomy. This allows the LLM to interpret multi-hop paths using explicit semantic rules.
For example, the system knows to rank a GWAS-derived association higher than a simple text-mined co-mention. It follows strict protocols for handling conflicting data or missing links. By constraining the model to these rules at runtime, the reasoning becomes independent of the underlying LLM’s "vibes."
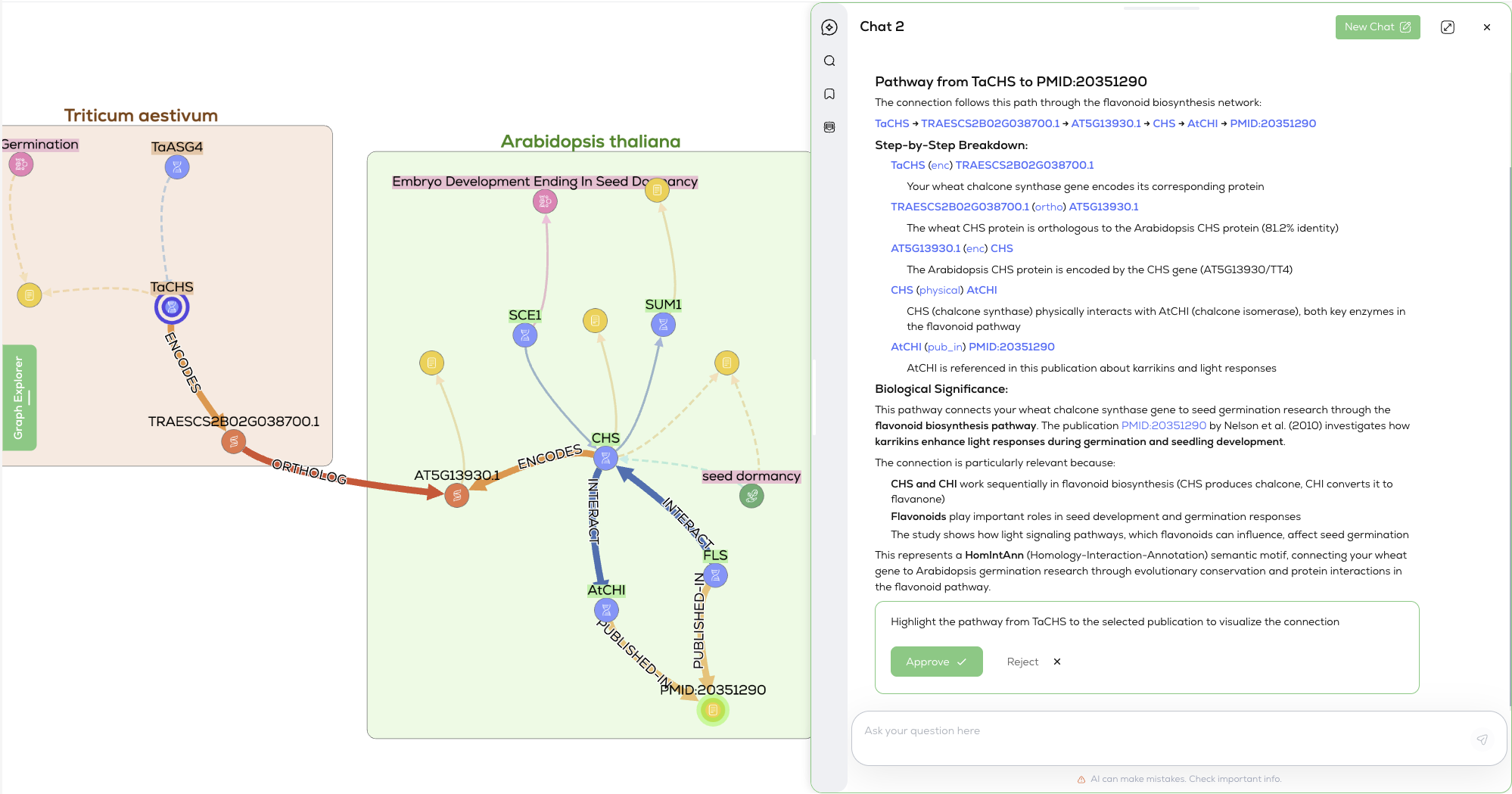
Navigable Biological Arguments
When you ask Graph Chat a question, you don’t just get a paragraph of text. You get a navigable biological argument. The response includes textual summaries cross-referenced to an interactive network.
The model understands the underlying logic of the path:
(gene:Gene) - [:enc] -> (:Protein) - [:ortho] - (:Protein) <- [:enc] - (:Gene) - [:has_variation] -> (:SNP) - [:associated_with] -> (:Phenotype)
This tight coupling between language and structure adds an essential layer of visual reasoning. Users aren't presented with a detached explanation; they can see the evidence, filter relationships by type, or reshape the graph layout to highlight specific mechanisms.
The Future of AI-Assisted Discovery
As LLMs become faster and cheaper, "fluent" answers will become a commodity. What will differentiate truly useful scientific systems is accountability.
Scientific discovery cannot rely on probabilistic guesswork. Graph-aware LLMs, guided by efficient context engineering rather than heavy fine-tuning, provide a scalable path to AI-assisted discovery without sacrificing the scientific rigor that researchers demand.
You might also like
.png)
.jpg)

Graph Chat: A New Way to Explore and Understand Biological Knowledge Graphs with AI

The Future is Green: How AI in Biotech is Revolutionising Plant Genomics











